Neuralgap Drug Discovery
Genesys Model Family
Architecturally designed for transparency. Natively multimodal. Radically efficient at atomic-level predictions.
Multi-Unimodal Architecture
Seamless integration across proteins, peptides, small molecules, and ligand analogs
Built-In Interpretability Layers: Genesys embeds algorithmic transparency tools – CAMs (Class Activation Maps), UMAPs, and edge detection – directly into its architecture. These expose how predictions are made at each layer, enabling atomic-level confidence scoring.
Architectural Efficiency (MUM + SOM): Our Multi-Unimodal framework and proprietary training techniques enable adaptive multimodal learning with less training data, lower compute costs, and better out-of-sample distribution performance.
Physics-Informed Design: Integrates structural constraints and conformational dynamics and thermodynamic endpoint approximations.
The Multi-Unimodal Architecture
Redefining Molecular Intelligence
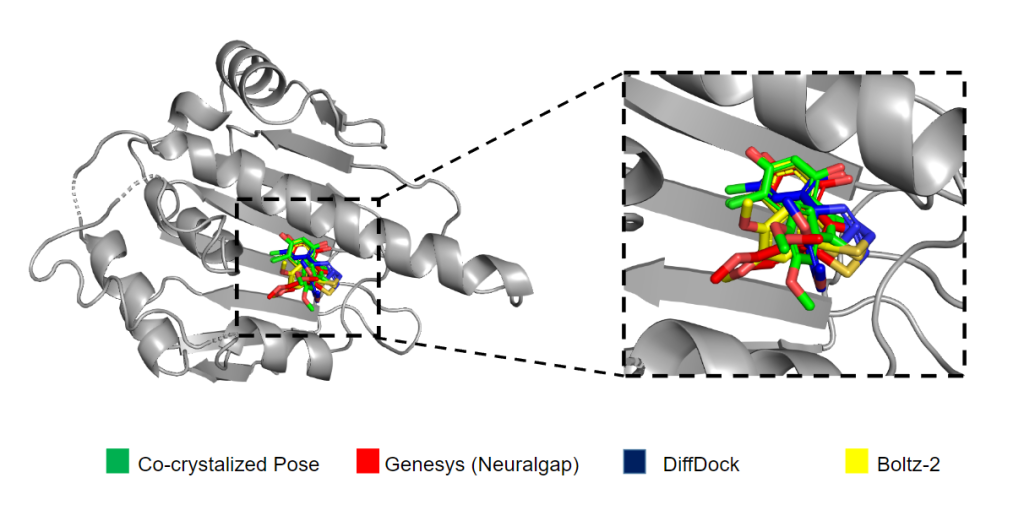
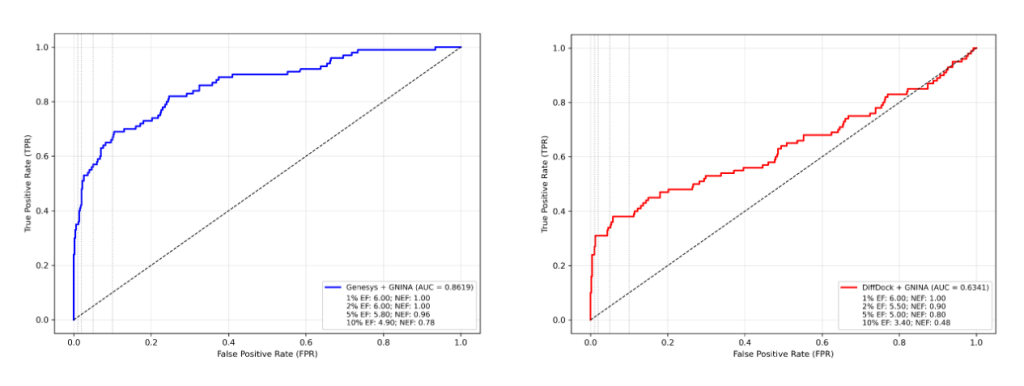
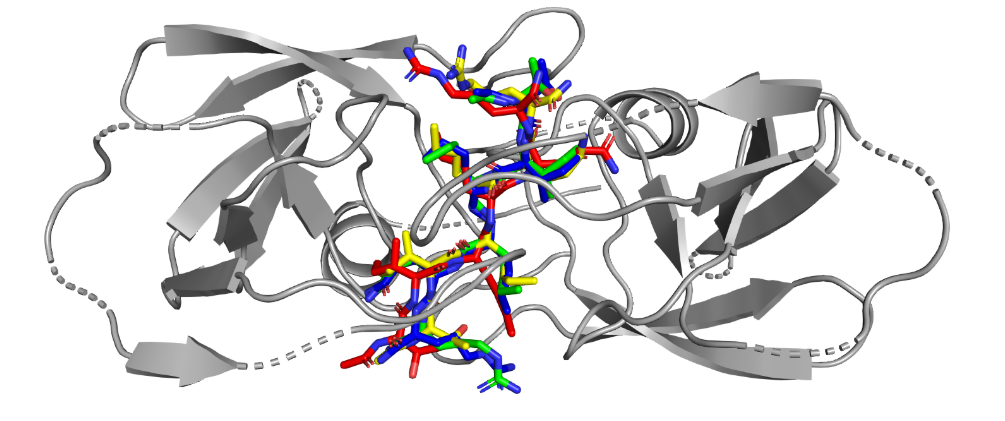
Multimodal by Architecture
- Native integration of proteins, peptides, small molecules, ligand analogs
- MUM framework enables incremental learning across modalities
- Self-organizing protein class selection
Efficient by Design
- 10x less training data than benchmarks
- Up to > 20x lower compute budgets
- Superior out-of-distribution generalization
Interpretable by Default
- Built-in transparency layers
- Native hooks for attention mapping, layer archaeology, conformational analysis
- Seamless integration with Circe’s confidence cartography
